

Sharma P, Wadhwa A, Komal K (2014) Analysis of selection schemes for solving an optimization problem in genetic algorithm.
#NONMEM ACCP SERIES#
Springer series in statistics (perspectives in statistics). In: Parzen E, Tanabe K, Kitagawa G (eds) Selected papers of Hirotugu Akaike. J Pharmacokinet Pharmacodyn 39(4):393–414Īkaike H (1998) Information theory and an extension of the maximum likelihood principle. Sherer EA, Sale ME, Pollock BG et al (2012) Application of a single-objective, hybrid genetic algorithm approach to pharmacokinetic model building. Icon Development Solutions, Ellicott City AAPS PharmSci 2(3):E32īeal S, Sheiner LB, Boeckmann A, Bauer RJ (2009) NONMEM user’s guides (1989–2009). Jonsson EN, Wade JR, Karlsson MO (2000) Nonlinearity detection: advantages of nonlinear mixed-effects modeling. Mitchell M (1996) An introduction to genetic algorithms. Goldberg DE (1989) Genetic algorithms in search, optimization, and machine learning. Wade JR, Beal SL, Sambol NC (1994) Interaction between structural, statistical, and covariate models in population pharmacokinetic analysis. Sale M, Sherer EA (2015) A genetic algorithm based global search strategy for population pharmacokinetic/pharmacodynamic model selection. This approach will further facilitate the scientist to shift efforts to focus on model evaluation, hypotheses generation, and interpretation and applications of resulting models. In addition to the GA capabilities, the workbench supports modeling efforts by: (1) Organizing and displaying models in tabular format, allowing the user to sort, filter, edit, create, and delete models seamlessly, (2) displaying run results, parameter estimates and precisions, (3) integrating xpose4 and PsN to facilitate generation of model diagnostic plots and run PsN scripts, (4) running regression models between post-hoc parameter estimates and covariates. A genetic algorithm implemented in an R-based NONMEM workbench for identification of near optimal models is presented. GAs are general, powerful, robust algorithms and can be used to find global optimal solutions for difficult problems even in the presence of non-differentiable functions, as is the case in the discrete nature of including/excluding model components in search of the best performing mixed-effects PK/PD model. Genetic algorithms are a class of algorithms inspired by the mathematics of evolution. Within the discipline of numerical optimization it falls into the “local search” category.
#NONMEM ACCP MANUAL#
Further, the current approach requires significant manual and redundant model modifications that heavily lend themselves to automation.
#NONMEM ACCP FREE#
If you know of more resources that can be added to this list, please feel free to provide the information by way of comments.The current approach to selection of a population PK/PD model is inherently flawed as it fails to account for interactions between structural, covariate, and statistical parameters.
#NONMEM ACCP UPDATE#
Nonlinear mixed effect models: an overview and update ĭ. Resources by Nick Holford: These course materials are freely available with enough background information to start performing population PKPD analysis.ī.Resource Facility for Population Pharmacokinetics – When accessing this site, you will have to fill a short questionnaire before downloading or viewing the tutorials/ presentations.Ĭ. In addition to the above discussion group, I have learnt immensely from the following resources.Ī.

My main objective of this post was to share some resources that are available to learn model data using NONMEM. Modeling and simulation has now become an integral part of drug development process and is being used regularly to model preclinical/clinical data.
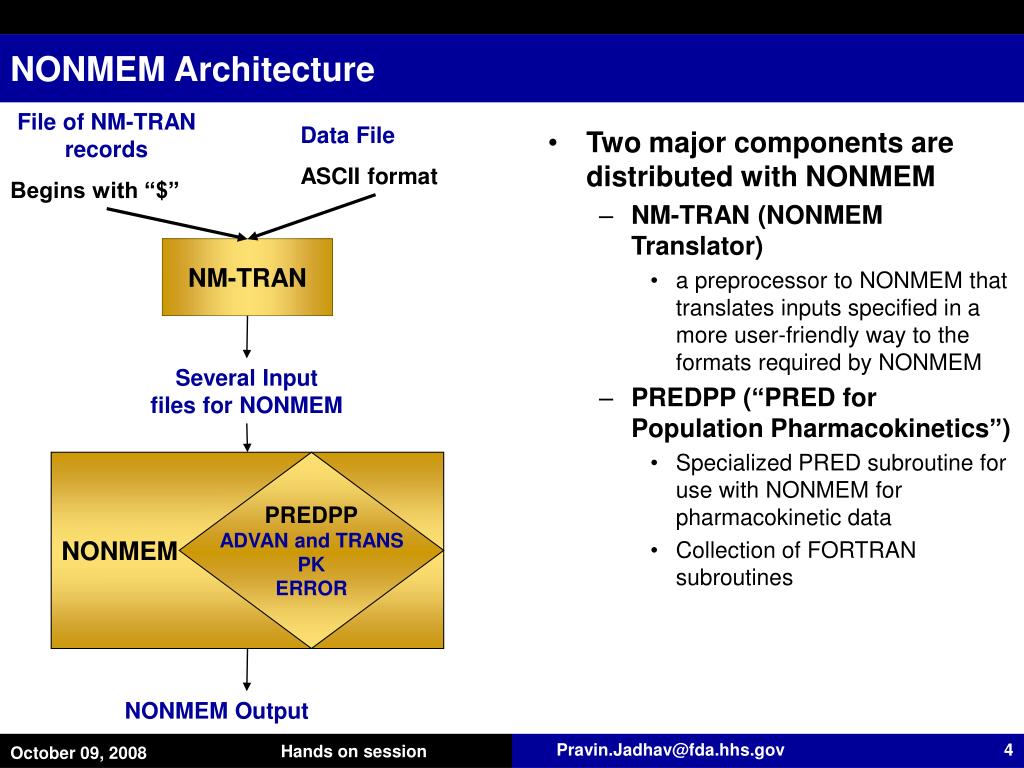
Roger Jeliffe MD, Jurgen Bulitta PhD about the use of NONMEM, USC Pack in population PK/PD modeling. We had guest lectures by Nick Holford MD, Dr. Each of us had an opportunity to use real clinical data obtained from paediatric patients and develop PK/PD models to describe the data. We have been meeting biweekly since 2006 and discuss about modeling data using NONMEM and other statistical softwares. The small group comprised of few MS/PhD graduate students (including me) and faculty from pharmacy/mathematics. I started learning NONMEM through a local discussion group headed by Dr.


 0 kommentar(er)
0 kommentar(er)
